time for some kind of anonymizing location data sharing service, peer to peer or federated protocol? that might be interesting, or sketchy, not sure which.
sixfold
Pretty sure you can download the maps ahead of time, GPS doesn't require data, then upload the fixes when you get home.
Well, it is technically differential equations, but with weighted inputs like a NN. Here's the equations


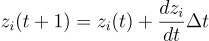
for each node (gene/morphogen) i. zi is the concentration of morphogen i, zj is that of j. f(x) is the sigmoid function, k1 is the maximum rate of expression, k2 is the degradation rate, b is the bias. wij is the weight for an edge from j to i.
This is just written in python, so the network is defined by a matrix with each number representing the weight between two of the edges. I ignore the edge if it's weight is zero.
What are the standard symbols for genetic circuits?
edit: sorry it's impossible to see the equations if you have a black background.
Damn, that's cool.
Here's the same 3 morphogen periodic clock network implemented in python https://diode.zone/w/9ApBpVcU5CnCbbTu3Lcegw

That's a super interesting project. For anyone else, the project overview has some great system level diagrams:
https://github.com/opentraffic/otv2-platform